Some Words about Cryo-Electron Tomography
Cryo-electron tomography (cryo-ET) is an imaging method used to obtain three-dimensional (3D) reconstructions of biological specimens in a close-to-native, vitrified state. Unlike single-particle analysis (SPA) – which aims to determine the structure of repeated, purified macromolecular complexes by averaging thousands or even millions of identical particles – cryo-ET focuses on imaging large and complex biological samples. These samples can be entire cells, subcellular organelles, or thin cellular sections (lamellae) produced by focused ion beam (FIB) milling.
The main difference between SPA and cryo-ET is that cryo-ET data is much more information-rich. In cryo-ET, you see much more than what you are specifically targeting, and sometimes it can even be difficult to find your target in the tomograms. Additionally, in cryo-ET, you typically work in 3D, even though the raw data are 2D projections of your sample. This means you need to examine your tomograms in detail to understand what’s in them, whereas in SPA, the micrographs often show the same structure multiple times.
Sample Types
Cryo-electron tomography (cryo-ET) requires thin samples (around 100–200 nm) so the electron beam of the transmission electron microscope (TEM) can pass through. For whole cells, a focused ion beam (FIB) is used to mill thin lamellae while preserving native structure. However, if you’re studying purified organelles (e.g., isolated mitochondria), you can sometimes skip or reduce milling if they’re already thin enough. People are also now going towards more complex samples (e.g small multicellular organisms or biopsies), which involves high-pressure freezing (HPF) of the sample instead of the more classical plunge-freezing.
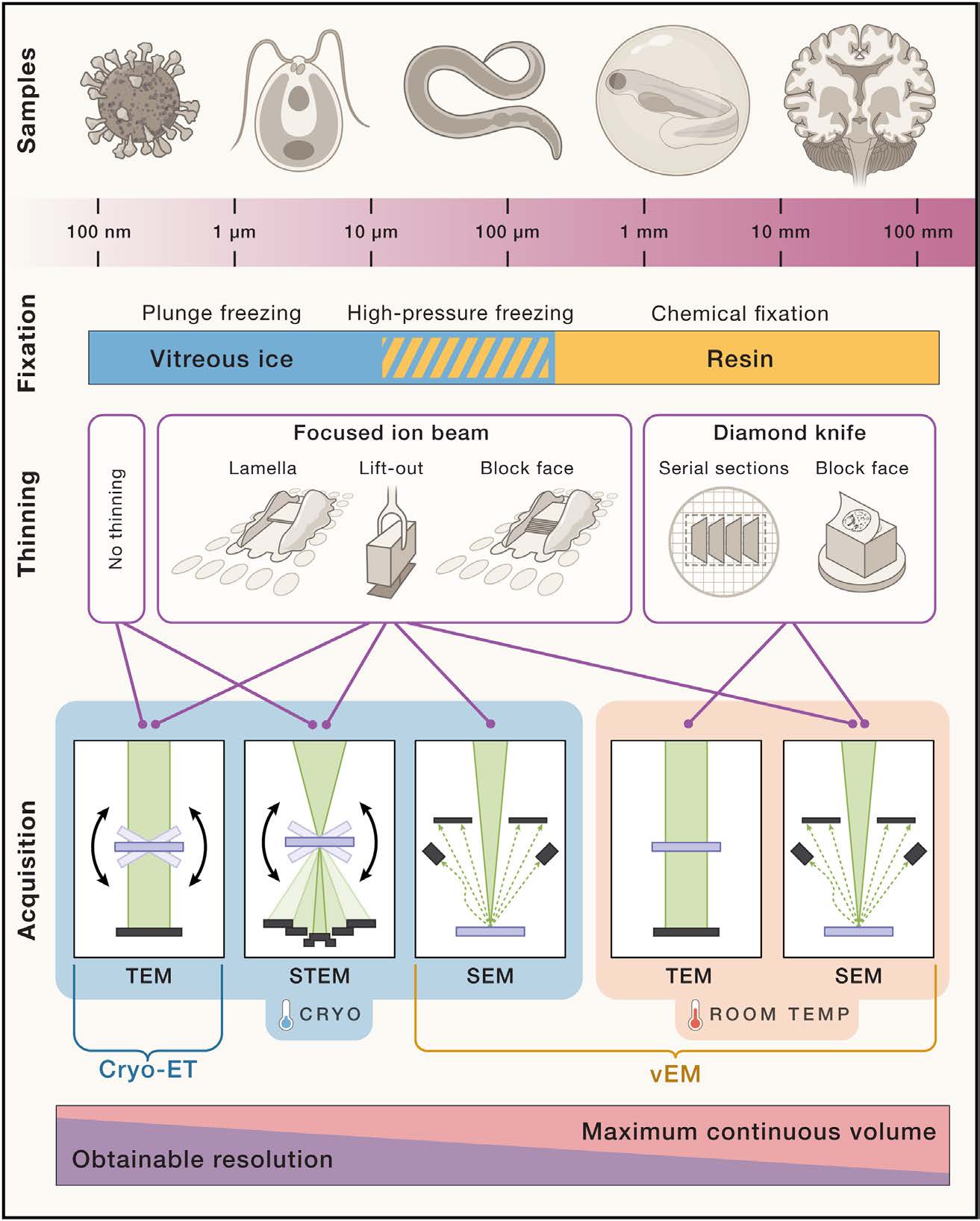
A critical philosophical difference is that cryo-ET aims to observe macromolecules in their natural cellular context. Meanwhile, SPA requires extensive purification steps, removing the protein complexes from their native milieu but achieving higher resolution in exchange for losing cellular context.
FIB-Milled Cellular Lamellae
- Preserves In Situ Context: You see organelles and macromolecules in their native environment.
- Complex and Heterogeneous: More challenging to prepare and interpret because you capture the full cellular complexity.
Purified sample (e.g organelles, viruses)
- Simpler Prep: Often no milling needed if the organelle is inherently small or can be fractionated to the right thickness.
- Less Context: You lose interactions with other cellular components—what you gain in simplicity and abundance, you lose in native spatial relationships.
In short, cellular lamellae capture the true cellular environment at the expense of more complex data collection and processing, whereas purified organelles simplify tomography but lose the broader context.
Cryo-ET Data Collection Workflow
-
Sample preparation
FIB-milled plunge-frozen cells, HPF samples, or purified macromolecules. -
Sample Mounting
The sample is transferred into the cryo-TEM (kept below –180°C). -
Tilt Series Acquisition
The stage is tilted incrementally (e.g., usually ±60° or ±40°) with 2–3° increments, collecting 2D projections at each tilt angle. -
Alignment
Projections are aligned to correct for stage shifts, beam-induced motion, etc. -
Tomogram Reconstruction
A 3D volume is reconstructed from aligned tilt series using weighted back-projection or SIRT. -
Post-processing & Further Analyses
You can segment specific structures in the tomogram or perform sub-tomogram averaging if you have repeated complexes.
Challenges in Cryo-ET
Dose and Radiation Damage
Electron radiation damages biological samples. The total electron dose must be strictly limited, leading to a lower signal-to-noise ratio (SNR).
Missing Wedge Artefact
Due to incomplete tilt sampling (typically ±60°), 3D reconstructions contain anisotropic information, known as the missing wedge effect.
Tilt-series alignment
Even small movements in the sample can degrade correlations between images.
Differences Between Cryo-ET and SPA
| Aspect | Cryo-ET | Single-Particle Analysis (SPA) |
|---|---|---|
| Sample Type | Unique cellular or subcellular specimens (e.g., FIB-milled lamellae or intact viruses/cells) | Purified, isolated macromolecular complexes or particles |
| 3D Reconstruction | 3D volume from multiple tilt images of the same specimen | 3D reconstruction from averaging many 2D projections of multiple copies of a particle in different orientations |
| Resolution | Typically lower (5–10 nm for cellular tomography, though can reach sub-nm with sub-tomogram averaging) | Can reach near-atomic resolutions (2–4 Å) for well-behaved particles and large data sets |
| Structural Context | In situ: preserves spatial relationships within the cell | In vitro: particles are studied in isolation, removed from their native environment |
| Data Requirement | Single tilt series per region of interest, capturing unique structures | Thousands to millions of particle images needed for high-resolution averaging |
| Throughput | Lower – time-consuming data collection per specimen | Potentially higher – automated data collection and well-optimized pipelines |