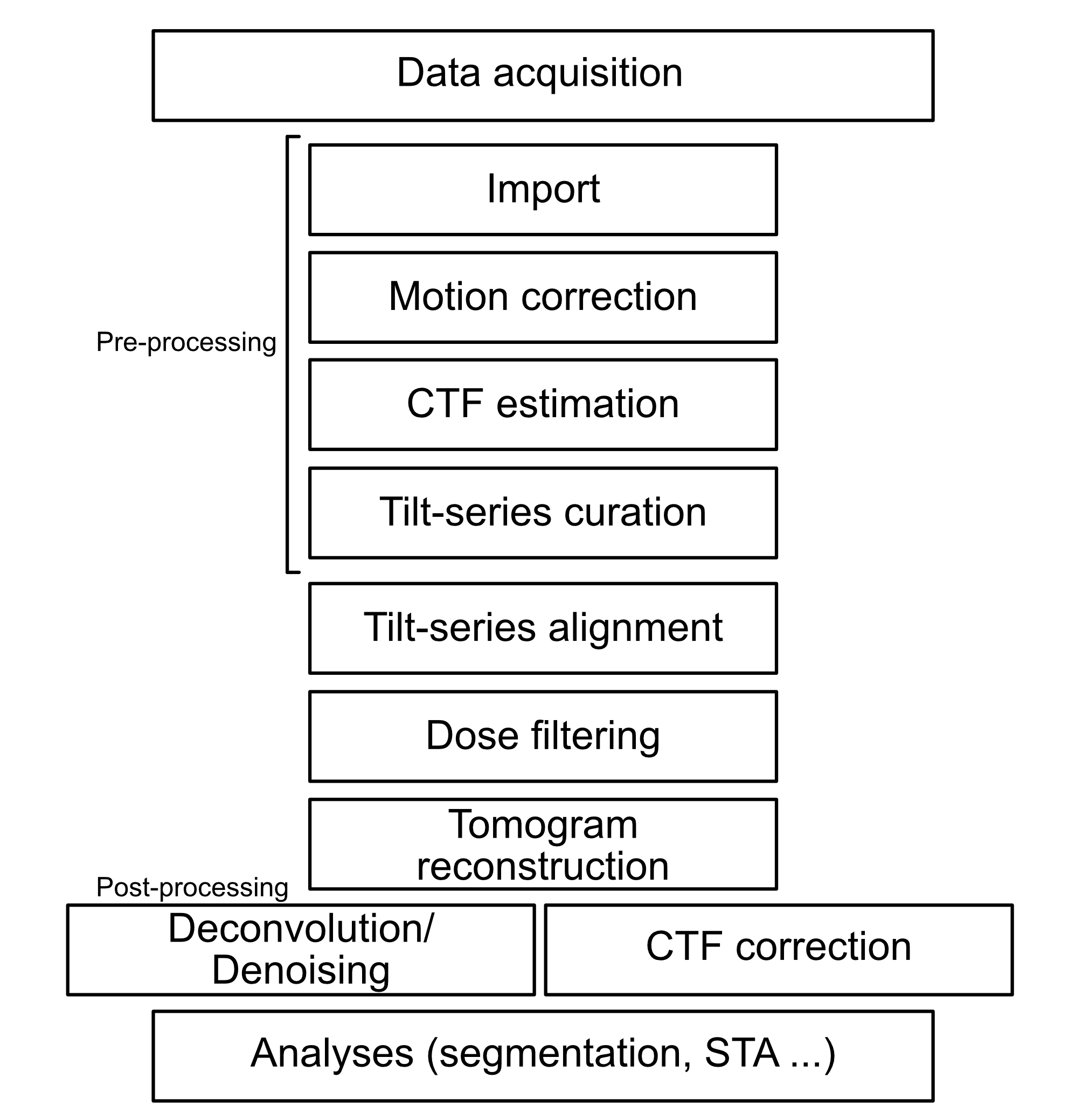
General workflow
Cryo-ET data processing can be summarised by this simple chart. If you’re familiar with single-particle data processing, you’ll notice that most of the pre-processing steps are very similar.
In this tutorial, we’ll walk through all the steps following data acquisition.
Here’s a quick summary:
Pre-processing
- Import: Load raw images into the analysis software and specify all the data acquisition parameters.
- Motion Correction: During imaging, the sample may drift slightly because it’s very sensitive to the electron beam. Motion correction detects and compensates for sample drifting caused by the electron beam, ensuring each image is as sharp and stable as possible.
- CTF Estimation: The electron microscope’s lenses can cause distortions in the images. The “Contrast Transfer Function” (CTF) describes those distortions. By estimating the CTF, you gather important information that helps correct these effects later on.
- Tilt-Series Curation: Because you take a series of images at various tilt angles, some may be of poor quality (e.g., too much drift or damage). Tilt-series curation is where you review the images and remove or flag the bad ones, ensuring only the best data moves forward.
Tilt-Series Alignment
Each tilted image is lined up properly with the others (kind of like stacking photos exactly on top of each other). Alignment makes sure that the features in each tilt angle match up in the correct positions to reconstruct the most accurate 3D volume.
Dose Filtering
High doses of electrons can damage the sample, and lower doses might lead to noisy images. Dose filtering involves adjusting images that are overly damaged or too low in signal to be useful. This helps maintain quality in the final 3D reconstruction.
Tomogram Reconstruction
Once all the tilt-series images are aligned and cleaned up, the software combines them to form a 3D “tomogram,” which is essentially a 3D picture of your sample.
Post-processing
- Deconvolution / Denoising: Specialised methods refine the 3D data by enhancing details and reducing noise, similar to using a ‘sharpen’ or ‘denoise’ filter on a standard image, only adapted for three dimensions. This is used to make the tomograms pleasant to “human” eyes.
- CTF Correction: You use the previously estimated CTF information to correct for the lens distortions in your final 3D tomogram, improving the overall quality and clarity. This is especially important for STA and automated particle picking.
Software
You need to have access to a GPU-powered machine running on Linux. It can be a local machine or a computing cluster. In our case, we work on a computing cluster with a SLURM system. You will also need to have appropriate CUDA drivers (this means you need to have NVIDIA GPUs) and a Python installation.
Click on the buttons below to get more information about the main software used in that tutorial and download it:
Scipion
IMOD
RELION 5
AreTomo3
ChimeraX
ArtiaX
pytom-match-pick