Check physical handedness of your tomograms
An essential aspect to consider when analyzing tomograms is to make sure they have the proper physical handedness.
Since tomograms are reconstructed from 2D projections of 3D objects, the third dimension’s orientation can be ambiguous, leading to potential mirror-image (left vs. right) inversions. This is also fairly common in SPA. This inversion is often imperceptible when viewing the tomogram, unless the sample contains distinct chiral features that are easily identifiable, such as the helical structure (e.g microtubules or cilia).
To determine the correct handedness, one effective method is to obtain an average of a structure within your tomogram (preferably non-symmetric). A good target, because they are large, abundant and easy to pick, are ribosomes.
Here are the structures of the same ribosome, one with the wrong and one with the correct handedness:
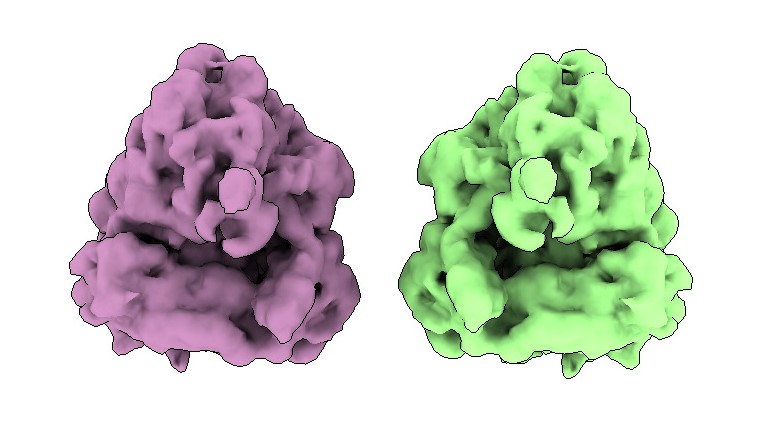
You can easily change the handedness of your volume in ChimeraX using the command:
vop flip #volume
At high resolution, you will also notice that DNA, RNA or alpha helices in proteins have the wrong pitch.
Getting the handedness wrong can easily happen if you use the wrong tilt axis at the import step. One easy fix if you notice that the handedness is wrong, is to change the sign of the tilt axis used (or more precisely, rotate by 180°).
Checking the handedness of your tomograms
If you are unsure of the handedness of your tomograms, the best is to template match 3-4 tomograms that contain ribosomes, with both a correct and a flipped template (example templates here).
Once you have run template matching, you should already see a difference by letting pytom determine the threshold for particle selection. One will output way more particles than the other.
Note that template matching with a template which has a different handedness than your tomo might still give you somewhat plausible results and fool you. Example:
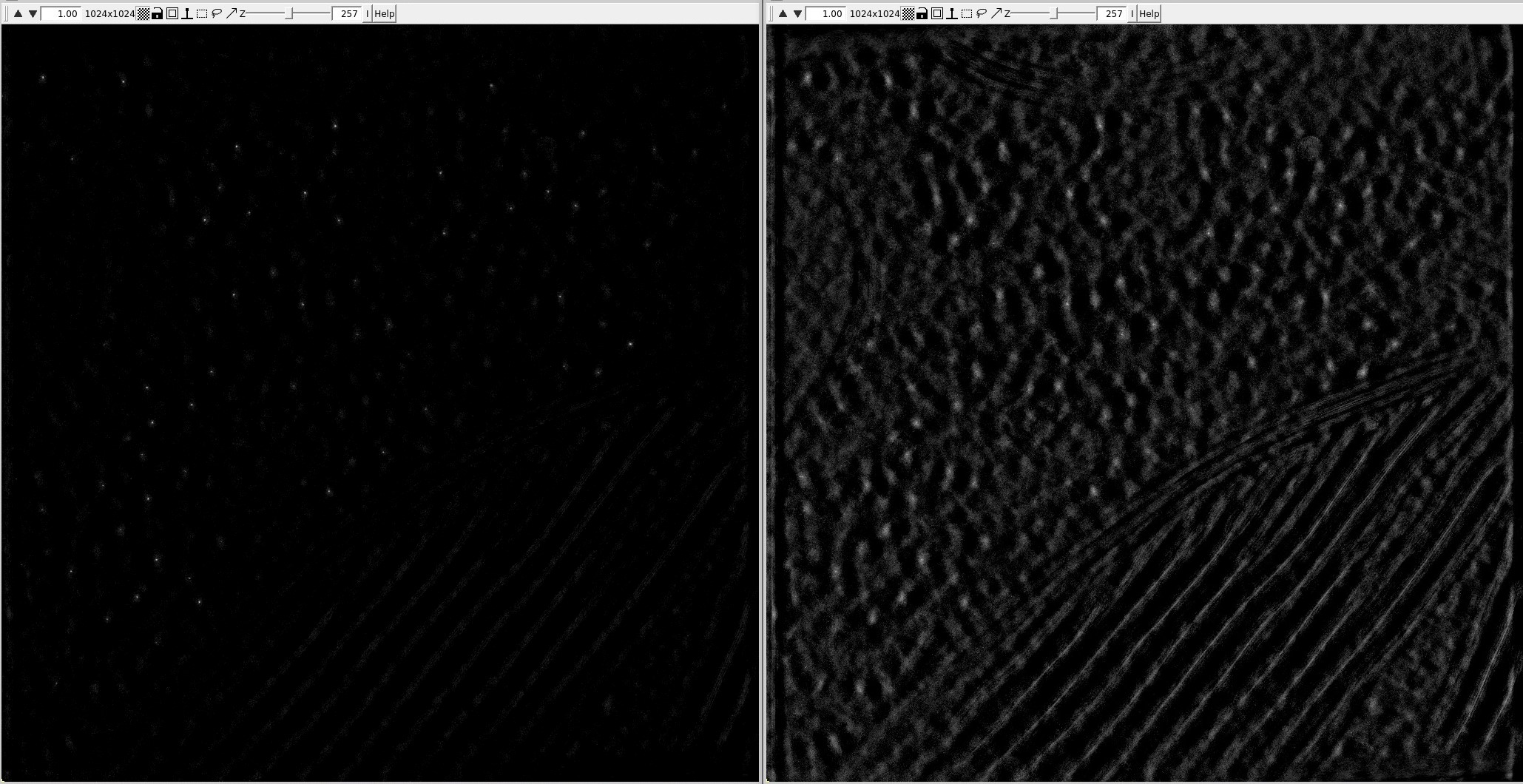
Here, the results of TM are shown using a proper template (left) or a flipped template (right).
The TM results are much better using the proper template (perfect sharp peaks), which means that the tomo has the proper hand, but the one on the right might look plausible to an inexperienced user.
Note that, for some reason, if operations were made on the microscope camera, this can also affect the handedness. So we would recommend double-checking the handedness if some operations were done on your microscope.
The handedness should, however, be the same for tilt-series acquired during the same sessions.
Defocus handedness
Note that there is another type of “handedness” to consider, the defocus handedness.
The defocus handedness describes whether defocus increases or decreases with Z height in a tomogram. It is not necessarily linked to the physical handedness concept described above.
Getting the defocus handedness wrong will not be noticeable until you reach high resolution in STA. Hence, it’s a bit less severe if you get this wrong.
In RELION5, this is specified by the “Invert defocus handedness” at the import step. You can check the defocus handedness using Defocusgrad, developed by Ricardo.